HDBSCAN clustering for identification of states with lignin dimers bound to β-cyclodextrin from molecular dynamics simulations
Introduction
Previously, we studied the interaction of β-cyclodextrin and lignin dimer derivatives using experimental methods, molecular dynamics simulations, and docking.[1] Chemical structures are shown in Figure 1. An introduction to the project is here, and a summary of the publication is here. Multiple bound states were observed in the molecular dynamics simulations. We estimated the proportions of those states in unbiased simulations using the following procedure:
- Computed a large number of collective variables including angle between lignin dimer and β-cyclodextrin principal axes, distances between atoms in the β-cyclodextrin molecule to atoms in the lignin dimer molecule, and lignin dihedral angles
- Used principal component analysis to reduce the number of dimensions to 2
- Used DBSCAN clustering to separate the different states into distinct clusters
- Counted the number of points in each cluster and computed proportions of each state
The details of the procedure are in the Supporting Info for [1] on pages 9-18. The main difficulty was in step 3. The states were not well separated and a lot of trial and error was required to choose DBSCAN parameters which gave separate clusters for them.
Here, only three collective variables were carefully chosen to separate the observed states. Therefore, dimensionality reduction was not required. Additionally, HDBSCAN was used instead of DBSCAN for clustering. Finding the first significant plateau in a plot of the number of clusters as a function of the min_samples = min_cluster_size parameter for HDBSCAN provided a way to identify a "natural" number of clusters. The final clusters were well separated. The analysis revealed a few states that were not apparent based on just visual inspection of the configurations in the trajectories.
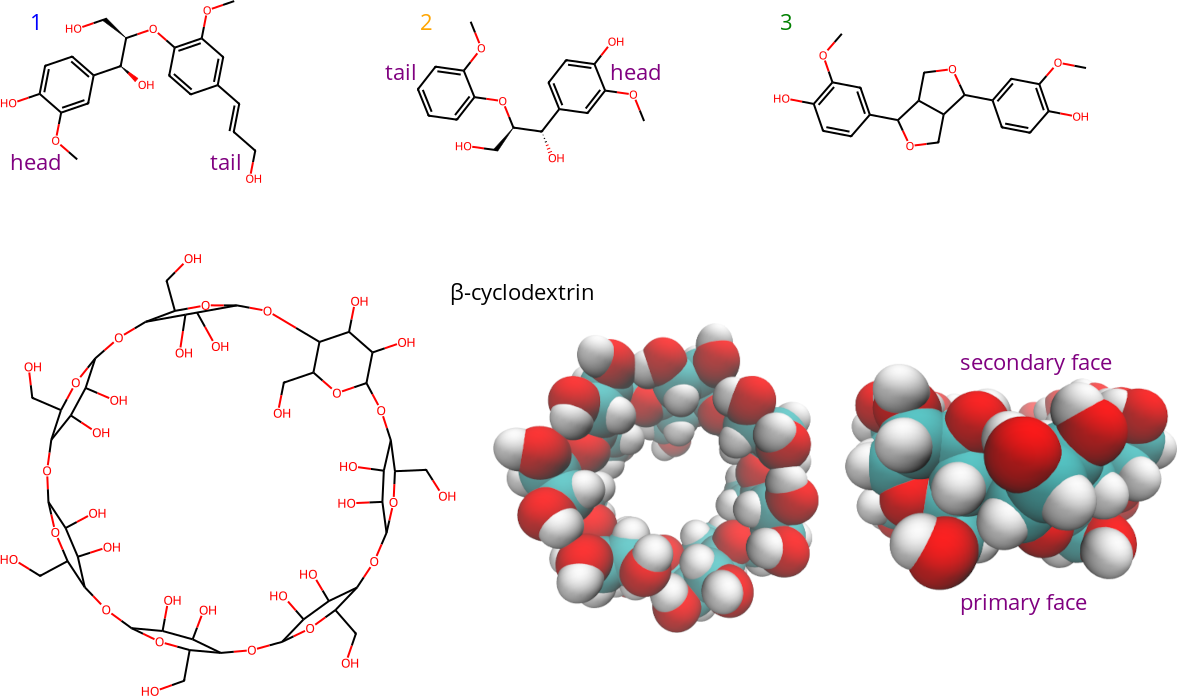
Figure 1: Structures of lignin dimer derivatives (1-3), and β-cyclodextrin. The lower right structures are 3D top and side views of β-cyclodextrin. The 3D representations show β-cyclodextrin from top and side views, with hydrogen atoms in white, carbon atoms in cyan, and oxygen atoms in red. The face of β-cyclodextrin with two hydroxyl (-OH) groups per unit (seen in the top view) is referred to as the secondary face, while the other face is referred to as the primary face.
Methods
Collective variables
Three collective variables were defined based on important configurations seen in unbiased simulations. The first was a signed distance along the direction defined by the vector pointing from the cyclodextrin center to the center of the oxygen atoms in the cyclodextrin secondary face.
The third collective variable was the cosine of the angle between the vector from the center of the atoms in the lignin head to the center of the atoms in the lignin tail (
The collective variables were computed using PLUMED driver version 2.6.
References
[1] Dean, K. R.; Novak, B.; Moradipour, M.; Tong, X.; Moldovan, D.; Knutson, B. L.; Rankin, S. E.; Lynn, B. C. Complexation of Lignin Dimers with β-Cyclodextrin and Binding Stability Analysis by ESI-MS, Isothermal Titration Calorimetry, and Molecular Dynamics Simulations. J. Phys. Chem. B 2022, 126 (8), 1655–1667.