Complexation of Lignin Dimers with β-Cyclodextrin and Binding Stability Analysis by ESI-MS, Isothermal Titration Calorimetry, and Molecular Dynamics Simulations
Introduction
Electrospray ionization mass spectrometry (ESI-MS), isothermal titration calorimetry (ITC), molecular dynamics (MD) simulations, and docking calculations were employed to investigate β-cyclodextrin interactions with lignin dimer derivatives. See Figure 1 for the chemical structures.
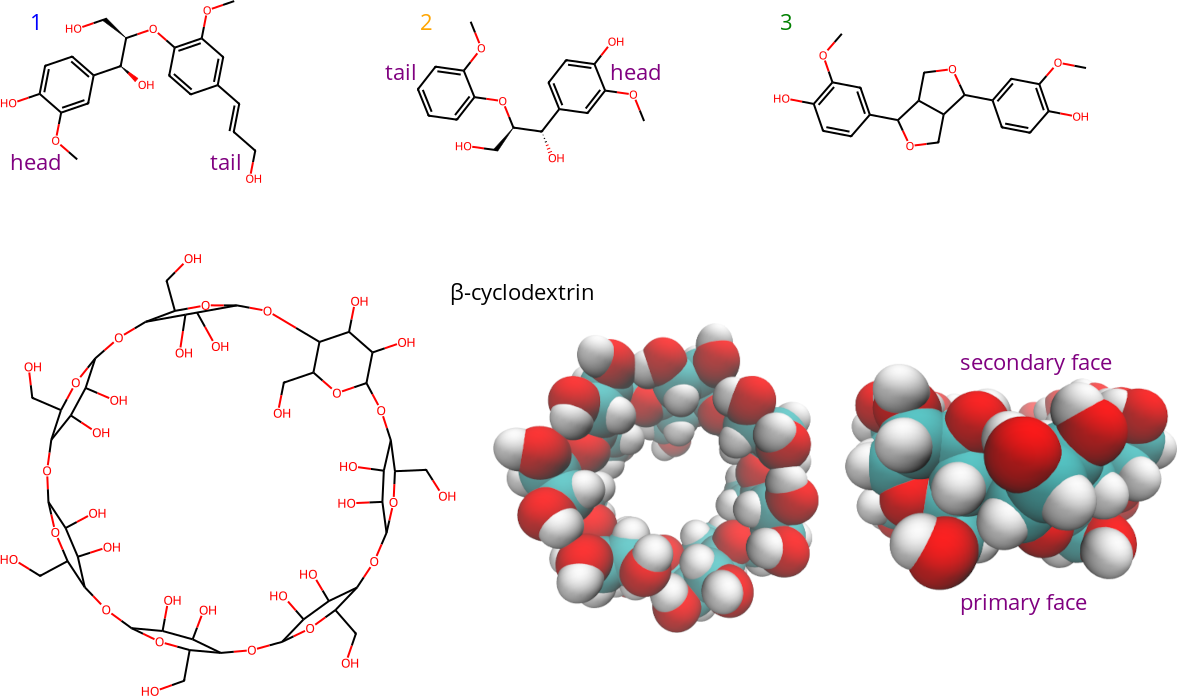
Figure 1: Structures of lignin dimer derivatives (1-3), and β-cyclodextrin. The lower right structures are 3D top and side views of β-cyclodextrin. The 3D representations show β-cyclodextrin from top and side views, with hydrogen atoms in white, carbon atoms in cyan, and oxygen atoms in red. The face of β-cyclodextrin with two hydroxyl (-OH) groups per unit (seen in the top view) is referred to as the secondary face, while the other face is referred to as the primary face.
Results
Binding free energy
Results indicated that dimer 3 was the most energetically favorable β-cyclodextrin guest. Table 1 shows the binding free energy differences, with dimer 3 having the most negative (strongest binding) value across all methods. Dimer 1 and 2 had similar, less negative values (weaker binding) compared to dimer 3, with some variations between the methods.
Table 1: Binding free energy differences (kJ/mol). For dimers 1 and 2, the MD results are for only one of the four isomers.
| Dimer | ESI-MS | ITC | MD | docking |
|---|---|---|---|---|
| 1 | -23.6 | -21.7 | -25.5 | -23.6 |
| 2 | -24.4 | -20.5 | -26.6 | -23.8 |
| 3 | -24.7 | -24.0 | -27.1 | -27.2 |
Observed complexes
ESI-MS experiments and MD simulations showed that both 1:1 and 2:1 complexes of β-cyclodextrin:lignin dimers were present. The prevalence of 2:1 complexes was higher for dimer 3 compared to dimers 1 and 2 in both methods. Specifically, in MD simulations, approximately 12% of β-cyclodextrin:dimer 2 complexes were 2:1 while approximately 48% of β-cyclodextrin:dimer 3 complexes were 2:1. Figure 2 shows configurations of complexes from the MD simulations.
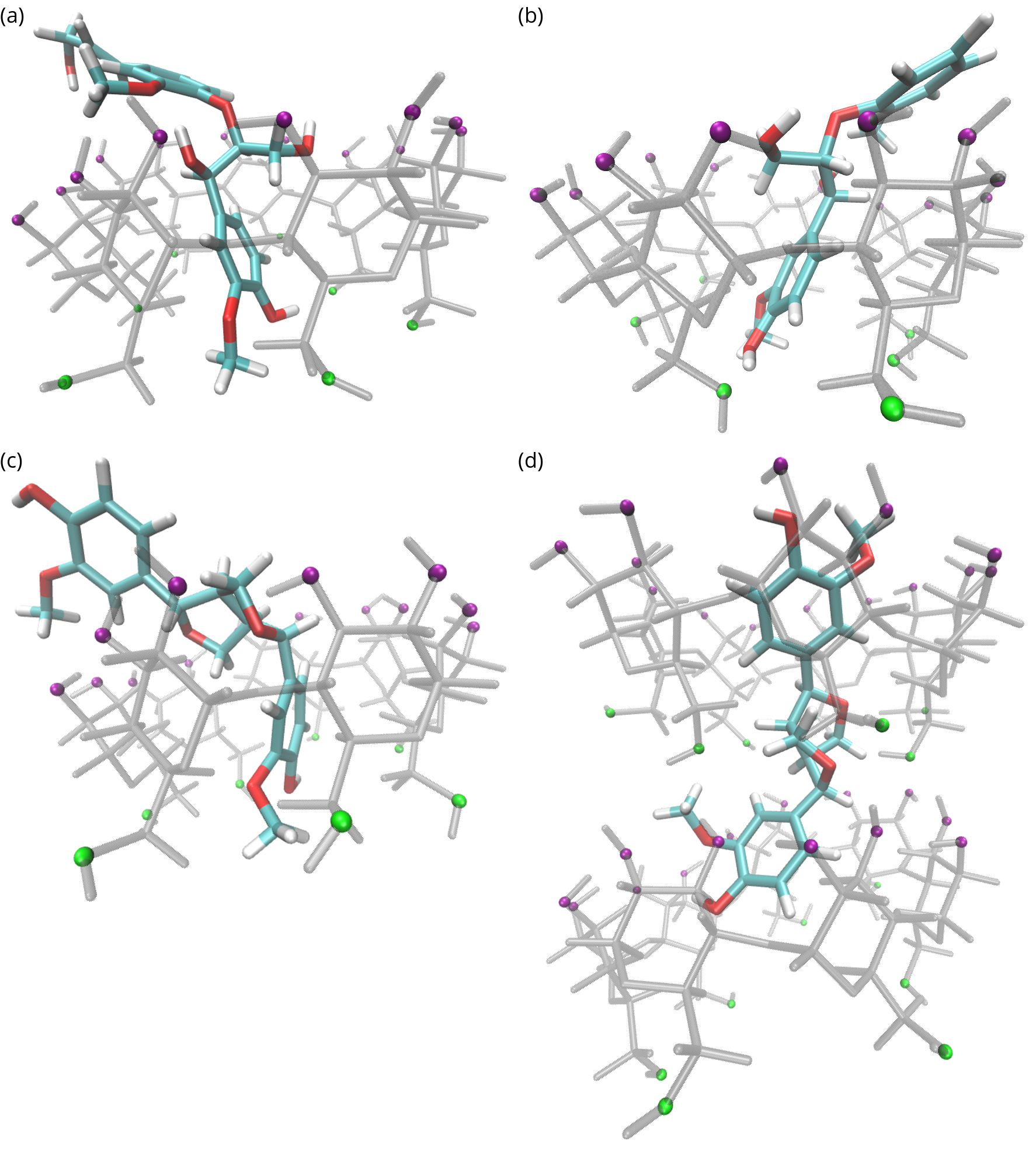
Figure 2: Dimer-cyclodextrin complexes. See Figure 1 for 2D structures and definitions. Cyclodextrin atoms are transparent grey with green primary face oxygen atoms and purple secondary face oxygen atoms. Lignin atoms are colored: hydrogen (white), oxygen (red), and carbon (cyan). (a) Dimer 1 bound headfirst through the cyclodextrin secondary face. (b) Dimer 2 bound headfirst through the cyclodextrin secondary face. (c) Dimer 3 bound through the cyclodextrin secondary face. (d) Dimer 3 bound to two cyclodextrin molecules.
The region connecting the two aromatic rings is more hydrophobic for dimer 3 compared to dimers 1 and 2 meaning it is more likely to be found at the center of the β-cyclodextrin which was observed in the MD simulations. Only one end of dimers 1 and 2 bound to the β-cyclodextrin center, but the central region of dimer 3 was frequently observed near the β-cyclodextrin center.
Binding animation
An interactive animation of dimer 1 binding to a β-cyclodextrin obtained from a MD simulation can be viewed here.